Protein-Protein Interactions in Solution and Crystallization
Introduction
 Interactions between
biomacromolecules in solution are a key factor determining the
phase behavior of biological systems. Also, the phase
behavior determines whether one can get good quality protein
crystals for X-ray diffraction, which is critical in obtaining
the proteins three-dimensional structure and further to
elucidate its biochemical role. [1-3] On the other hand, the
protein interaction and aggregation processes are also very
important in understanding many physiological problems, for
example, diseases like Alzheimer or Kreutzfeld-Jacob and
Parkinson, which are caused by protein or peptide association
phenomena. Among the various factors, the electrostatic
effects play a major role in protein structure, interactions,
and dynamics. They depend on the protein itself, the
surrounding water, and ions present (see detail). Interactions between
biomacromolecules in solution are a key factor determining the
phase behavior of biological systems. Also, the phase
behavior determines whether one can get good quality protein
crystals for X-ray diffraction, which is critical in obtaining
the proteins three-dimensional structure and further to
elucidate its biochemical role. [1-3] On the other hand, the
protein interaction and aggregation processes are also very
important in understanding many physiological problems, for
example, diseases like Alzheimer or Kreutzfeld-Jacob and
Parkinson, which are caused by protein or peptide association
phenomena. Among the various factors, the electrostatic
effects play a major role in protein structure, interactions,
and dynamics. They depend on the protein itself, the
surrounding water, and ions present (see detail).
George and Wilson [4] proposed a relation between protein
crystallization behavior and the osmotic second virial
coefficient, A2, which represents the interaction potential
between a pair of macromolecules in solution. A positive
value of A2 implies a repulsive interaction and a negative
value indicates an attractive interaction. Based on
measurements of a variety of proteins, they found that protein
crystallization occurs only when A2 lies within a narrow
window. These studies provide a way to understand the
mechanism of protein crystallization and a guide for
optimization of conditions for protein crystallization. [5-7]
On the other hand, the protein interaction and aggregation
processes are also very important in understanding many
physiological problems, for example, diseases like Alzheimer
or Kreutzfeld-Jacob and Parkinson, which are caused by protein
or peptide association phenomena, and the short-range order of
(-crystalline accounts for the eye lens transparency. [3,8] In
vivo, the biochemical function of proteins cannot perform
without the cooperation of ions around them. Therefore,
studies on the effect of ionic strength and the nature of ions
on the protein interaction have attracted much attention in
biophysics. [5,6,9] Studies show that the interaction strongly
depends on the nature of salt used at a fixed ionic strength
which is known as the "Hofmeister effect". [10]
This project addresses the important question of
- how
ions surrounding a protein interact with the protein and its
hydration shell
- how ions influence specific and unspecific protein-protein
interactions, and
- how ions influence flexibility and dynamics of proteins
and protein-complexes.
|
|
To answer these questions, the density and location of ions
around the protein, their specific interaction with surface
charges and the dynamics of these ions and the solvent will be
determined. These effects influence protein-protein
aggregation, the stability of protein-protein complexes, and
flexibility of individual protein side chains or whole loop
regions.
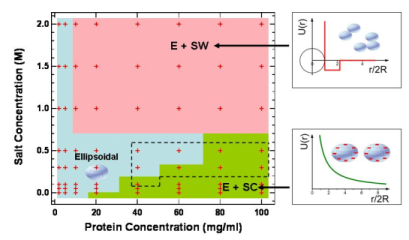
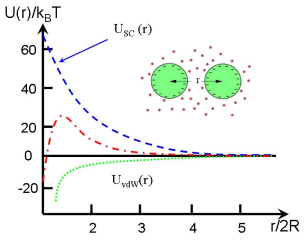
Typical interaction potential is described by DLVO theory
which includes a screened Coulomb repulsion with a hard sphere
core and an attraction of van der Waals interaction.
|
|
Experiments
|
By the combination of theoretical and experimental studies we gain
a comprehensive picture of charge effects in this context. [11,12] The
techniques complement each other, revealing a number of different
aspects of charge effects. Experimentally, optical
microscopy (view
real-time video) , spectroscopic
lab-based techniques (IR, UV/vis, CD) as well as neutron and
synchrotron-based scattering techniques (SAXS, ASAXS, QENS) are
employed to determine the structure, the salt-ion distribution, and
various aspects of the dynamics of individual proteins and protein
complexes. Theoretical studies based on local and nonlocal continuum
electrostatics calculations and molecular dynamics simulations
complement the spectroscopic techniques. These techniques reveal
counter ion distribution, the effect of ions on electrostatic
shielding and molecular recognition, counter ion mobility and the
influence of charge on local protein motion at atomic resolution. , spectroscopic
lab-based techniques (IR, UV/vis, CD) as well as neutron and
synchrotron-based scattering techniques (SAXS, ASAXS, QENS) are
employed to determine the structure, the salt-ion distribution, and
various aspects of the dynamics of individual proteins and protein
complexes. Theoretical studies based on local and nonlocal continuum
electrostatics calculations and molecular dynamics simulations
complement the spectroscopic techniques. These techniques reveal
counter ion distribution, the effect of ions on electrostatic
shielding and molecular recognition, counter ion mobility and the
influence of charge on local protein motion at atomic resolution.
|
Currently, we are interested in the following issues:
- Phase behaviour of model globular proteins in solution
with salt addition; Combined with liquid-state theoretical
approaches, the protein-protein interaction potentials can
be extracted from SAS data analysis.
- Multivalent counterion distribution around charged
proteins in solution studied by SAS and anomalous SAXS;
- Protein thermal aggregation studied by real time SAXS;
- Protein dynamics studied by neutron spectroscopy
[1] Durbin, S. D.; Feher, G. Annu. Rev. Phys. Chem. 1996, 47, 171.
[2] Anderson, V. J.; Lekkerkerker, H. N. W. Nature 2002, 416, 811.
[3] Piazza, R.; Curr. Opin. Colloid Interface Sci. 2004, 8, 515.
[4] George, A.; Wilson, W. Acta Cryst. 1994, D50, 361.
[5] Tardieu, A.; Le Verge, A.; Malfois, M.; Bonneté, F.; Finet, S.; Riès-
Kautt, M.; Belloni, L. J. Cryst. Growth 1999, 196, 193.
[6] Bonneté, F.; Finet, S.; Tardieu, A. J. Cryst. Growth 1999, 196, 403.
[7] Stradner, A.; Sedgwich, H.; Cardinaux, F.; Poon, W. C. K.; Egelhaaf, S.
U.; Schurtenberger, P. Nature 2004, 432, 492.
[8] Delaye, M.; Tardieu, A. Nature 1983, 302, 415.
[9] Curtis, R. A.; Prausnitz, J. M.; Blanch, H. W. Biotechnol. Bioeng. 1998,
57, 11.
[10] Collins, K. D. Biophys. J. 1997, 72, 65, Collins, K. D.; Washabaugh, M.
W. Q. Rev. Biophys. 1985, 18, 323
[11] F. Zhang, M.W.A. Skoda, R.M.J. Jacobs, R.A. Martin, C.M. Martin, F.
Schreiber, J. Phys. Chem. B, 111, 251 (2007)
[12] F. Zhang, M.W.A. Skoda, R.M.J. Jacobs, S. Zorn, R.A.
Martin, C.M. Martin, G. F. Clark, S. Weggler, A. Hildebrandt,
O. Kohlbacher, F. Schreiber, Phys. Rev. Lett. 101 (2008) 148101
For our recent work on proteins in solution,
see list of publications.
 Interactions between
biomacromolecules in solution are a key factor determining the
phase behavior of biological systems. Also, the phase
behavior determines whether one can get good quality protein
crystals for X-ray diffraction, which is critical in obtaining
the proteins three-dimensional structure and further to
elucidate its biochemical role. [1-3] On the other hand, the
protein interaction and aggregation processes are also very
important in understanding many physiological problems, for
example, diseases like Alzheimer or Kreutzfeld-Jacob and
Parkinson, which are caused by protein or peptide association
phenomena. Among the various factors, the electrostatic
effects play a major role in protein structure, interactions,
and dynamics. They depend on the protein itself, the
surrounding water, and ions present (see detail).
Interactions between
biomacromolecules in solution are a key factor determining the
phase behavior of biological systems. Also, the phase
behavior determines whether one can get good quality protein
crystals for X-ray diffraction, which is critical in obtaining
the proteins three-dimensional structure and further to
elucidate its biochemical role. [1-3] On the other hand, the
protein interaction and aggregation processes are also very
important in understanding many physiological problems, for
example, diseases like Alzheimer or Kreutzfeld-Jacob and
Parkinson, which are caused by protein or peptide association
phenomena. Among the various factors, the electrostatic
effects play a major role in protein structure, interactions,
and dynamics. They depend on the protein itself, the
surrounding water, and ions present (see detail).
 , spectroscopic
lab-based techniques (IR, UV/vis, CD) as well as neutron and
synchrotron-based scattering techniques (SAXS, ASAXS, QENS) are
employed to determine the structure, the salt-ion distribution, and
various aspects of the dynamics of individual proteins and protein
complexes. Theoretical studies based on local and nonlocal continuum
electrostatics calculations and molecular dynamics simulations
complement the spectroscopic techniques. These techniques reveal
counter ion distribution, the effect of ions on electrostatic
shielding and molecular recognition, counter ion mobility and the
influence of charge on local protein motion at atomic resolution.
, spectroscopic
lab-based techniques (IR, UV/vis, CD) as well as neutron and
synchrotron-based scattering techniques (SAXS, ASAXS, QENS) are
employed to determine the structure, the salt-ion distribution, and
various aspects of the dynamics of individual proteins and protein
complexes. Theoretical studies based on local and nonlocal continuum
electrostatics calculations and molecular dynamics simulations
complement the spectroscopic techniques. These techniques reveal
counter ion distribution, the effect of ions on electrostatic
shielding and molecular recognition, counter ion mobility and the
influence of charge on local protein motion at atomic resolution.