Small-Angle X-ray Scattering (SAXS)
The small-angle scattering technique can be used to study the
structure and interactions of colloidal solutions. Particularly
the properties of biological macromolecules such as DNA and
proteins, which may be regarded as (charged) colloids, can be
determined by SAXS measurements.
|
According to Bragg's law
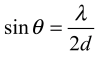
and for wavelengths λ around 1 Å the scattering
angle 2θ corresponding to atomic distances d of a few
Å is relatively large, i.e. typically 20°.
Since colloids, biological macromolecules such as polymers,
amphiphilic systems, membranes and liquid crystals exhibit
typical distances d or domain sizes of 100 Å, the
corresponding scattering angle 2θ is much smaller,
i.e. typically 0.5°. All structural information is contained
in a small angular range - which is why the technique is called
"small-angle scattering". |
|
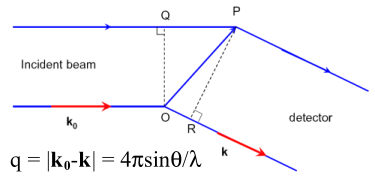
The total scattering intensity I(q) for a monodisperse
spherical system at a scattering angle 2θ as a function of
the scattering vector q can be expressed by:
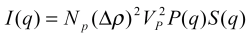
where
- NP is the number of protein molecules per
unit volume in the solution,
- VP is the volume of a single protein,
- Δρ = (ρp -
ρs), is the difference between the electron
density of protein molecules and that of the solvent
(usually called the scattering contrast),
- P(q) is the form factor of a given protein, i. e. the
scattering from a single protein molecule after orientational
averaging,
- S(q) is the structure factor, which contains information on
the protein interactions
For a polydisperse or non-spherical system S(q) may be
replaced by an effective structure factor, which is calculated
using a monodisperse structure factor at an effective sphere
diameter. Further details on small-angle scattering and the data
analysis can be found in this
tutorial.
We have a lab-based small-angle X-ray scattering
setup
(Details) .
.
Moreover, we have access to small-angle beamlines at different
synchrotron radiation sources, such as the ID02 at the ESRF.
We recently studied interactions of a model protein, bovine
serum albumin (BSA) in aqueous solutions as a function of protein
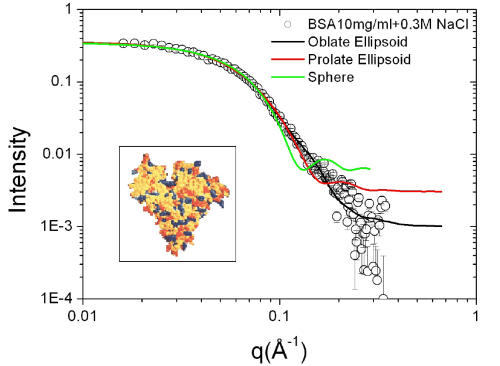
concentration and ionic strength. First, the form factor of
single protein molecule can be determined from the scattering of
dilute protein solution with moderate ionic strength as shown in
Figure 3. An oblate ellipsoidal form factor gives the best fit of
the scattering int ensity. BSA molecules at pH 7 are negatively
charged, therefore, at low ionic st rength, the interactions
between proteins are dominated by electrostatic
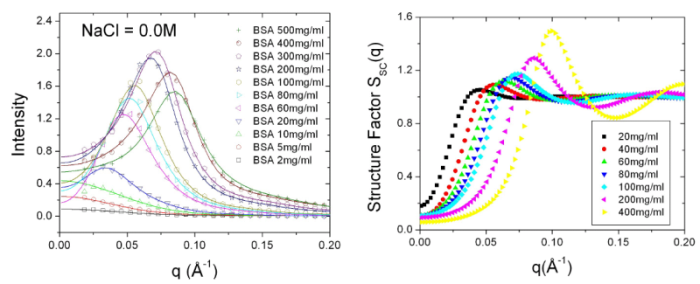
repulsion. Combined with screened Coulomb structure factor, the
scattering profiles from moderate to high protein concentration
solution can be well-fitted and the corresponding structure
factors are also evaluated from data fitting.
References
[1] R. J. Roe, "Methods of X-ray and Neutron Scattering in Polymer
Science", Oxford, Oxford University Press, 2000.
[2] P. Lindner, Th. Zemb, Ed. "Neutrons, X-rays and Light:
Scattering Methods Applied to Soft Condensed Matter",
North-Holland, Elsevier, 2002.
[3] F. Zhang, M. Skoda, R. Jacobs, R. A. Martin, C. M. Martin,
F. Schreiber, J. Phys. Chem. B 2007, 111, 251-259.
For our previous work on SAXS, see our list
of publications.
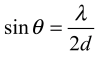
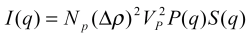
 .
.